Getting started tutorial part 3: displaying exposures and source tables output by processCcd.py¶
In the previous tutorial in the series you used processCcd.py to calibrate a set of raw Hyper Suprime-Cam images. Now you’ll learn how to use the LSST Science Pipelines to inspect processCcd.py’s outputs by displaying images and source catalogs in the DS9 image viewer. In doing so, you’ll be introduced to some of the LSST Science Pipelines’ Python APIs, including:
- Accessing datasets with the Butler.
- Displaying images in DS9 with
lsst.afw.display. - Working with source catalogs using
lsst.afw.table.
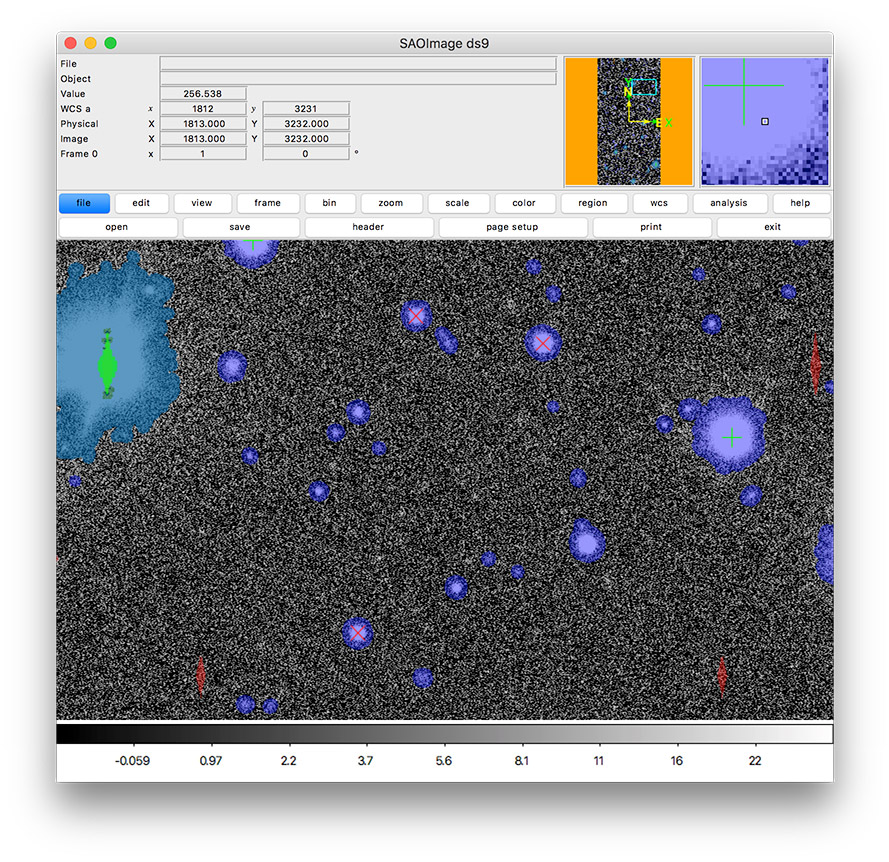
In this tutorial, you’ll create an image display like this one that includes mask planes and source markers.
Set up¶
Pick up your shell session where you left off in part 2.
That means your current working directory must contain the DATA directory (the Butler repository).
The lsst_distrib package also needs to be set up in your shell environment.
See Setting up installed LSST Science Pipelines for details on doing this.
You’ll also need to download and install the DS9 image viewer.
Launch DS9 and start a Python interpreter¶
In this tutorial, you will use an interactive Python session to control DS9.
If you haven’t already, launch the DS9 application.
Next, start up a Python interpreter. You can use the default Python shell (python), the IPython shell, or even run from a Jupyter Notebook. Ensure that this Python session is running from the shell where you ran setup lsst_distrib.
Creating a Butler client¶
All data in the Pipelines flows through the Butler.
As you saw in the previous tutorial, processCcd.py read exposures from the Butler repository and persisted outputs back to the repository.
Although this Butler data repository is a directory on the filesystem (DATA), we don’t recommend directly accessing its files.
Instead, you use the Butler client from the lsst.daf.persistence module.
In the Python interpreter, run:
import lsst.daf.persistence as dafPersist
butler = dafPersist.Butler(inputs='DATA/rerun/processCcdOutputs')
The Butler client reads from the data repository specified with the inputs argument.
In the previous tutorial, you created the processCcdOutputs rerun to isolate the outputs of the processCcd.py command-line task.
Reruns act like repositories, so to work with the processCcd.py outputs you specifically set inputs as the path to that rerun.
Tip
Reruns are sub-directories of the rerun directory of a root Butler data repository.
Listing available data IDs in the Butler¶
To get data from the Butler you need to know two things: the dataset type and the data ID.
Every dataset stored by the Butler has a well-defined type.
Tasks read specific dataset types and output other specific dataset types.
The processCcd.py command reads in raw datasets and outputs calexp, or calibrated exposure, datasets (among others).
It’s calexp datasets that you’ll display in this tutorial.
Data IDs let you reference specific instances of a dataset.
On the command line you select data IDs with --id arguments, filtering by keys like visit, ccd, and filter.
Now, use the Butler client to find what data IDs are available for the calexp dataset type:
butler.queryMetadata('calexp', ['visit', 'ccd'], dataId={'filter': 'HSC-R'})
The printed output is a list of (visit, ccd) key tuples for all data IDs where the filter key is the HSC-R band:
[(903334, 16),
(903334, 22),
(903334, 23),
(903334, 100),
(903336, 17),
(903336, 24),
(903338, 18),
(903338, 25),
(903342, 4),
(903342, 10),
(903342, 100),
(903344, 0),
(903344, 5),
(903344, 11),
(903346, 1),
(903346, 6),
(903346, 12)]
Note
That example butler.queryMetadata call is equivalent to this shell command that you used in the previous tutorial:
processCcd.py DATA --rerun processCcdOutputs --id filter=HSC-R --show data
Get an exposure through the Butler¶
Knowing a specific data ID, let’s get the dataset with the Butler client’s get method:
calexp = butler.get('calexp', dataId={'filter': 'HSC-R', 'visit': 903334, 'ccd': 23})
This calexp is an ExposureF Python object.
Exposures are powerful representations of image data because they contain not only the image data, but also a variance image for uncertainty propagation, a bit mask image plane, and key-value metadata.
In the next steps you’ll learn how to display an Exposure’s image and mask.
Create a display¶
To display the calexp you will use the display framework, which is imported as:
import lsst.afw.display as afwDisplay
The display framework provides a uniform API for multiple display backends, including DS9 and LSST’s Firefly viewer.
For this tutorial you’ll create a display with the ds9 backend:
display = afwDisplay.getDisplay(backend='ds9')
Display the calexp (calibrated exposure)¶
Then use the display’s mtv method to view the calexp in DS9:
display.mtv(calexp)
As soon as you execute the command a single Hyper Suprime-Cam calibrated exposure, the {'filter': 'HSC-R', 'visit': 903334, 'ccd': 23} data ID, should appear in the DS9 application.
Notice that the DS9 display is filled with colorful regions. These are mask regions. Each color reflects a different mask bit that correspond to detections and different types of detector artifacts. You’ll learn how to interpret these colors later, but first you’ll likely want to adjust the image display.
Improving the image display¶
The display framework gives you control over the image display to help bring out image details.
To make masked regions semi-transparent, so that underlying image features are visible, try:
display.setMaskTransparency(60)
The setMaskTransparency method’s argument can range from 0 (fully opaque) to 100 (fully transparent).
You can also control the colorbar scaling algorithm with the display’s scale method.
Try an asinh stretch with the zscale algorithm for automatically selecting the white and black thresholds:
display.scale("asinh", "zscale")
Instead of an automatic algorithm like zscale (or minmax) you can explicitly provide both a minimum (black) and maximum (white) value:
display.scale("asinh", -1, 30)
Interpreting displayed mask colors¶
The display framework renders each plane of the mask in a different color (plane being a different bit in the mask).
To interpret these colors you can get a dictionary of mask planes from the calexp and query the display for the colors it rendered each mask plane with.
Run:
mask = calexp.getMask()
for maskName, maskBit in mask.getMaskPlaneDict().items():
print('{}: {}'.format(maskName, display.getMaskPlaneColor(maskName)))
As an example, this result is:
DETECTED_NEGATIVE: cyan
CROSSTALK: None
INTRP: green
DETECTED: blue
UNMASKEDNAN: None
NO_DATA: orange
BAD: red
EDGE: yellow
SUSPECT: yellow
NOT_DEBLENDED: None
CR: magenta
SAT: green
Footprints of detected sources are rendered in blue and the saturated cores of bright stars are drawn in green.
Tip
Try customizing the color of a mask plane with the Display.setMaskPlaneColor method.
You can choose any X11 color.
For example:
display.setMaskPlaneColor('DETECTED', 'dodgerblue')
display.mtv(calexp)
Getting the source catalog generated by processCcd.py¶
Besides the calibrated exposure (calexp), processCcd.py also creates a table of the sources it used for PSF estimation as well as astrometric and photometric calibration.
The dataset type of this table is src, which you can get from the Butler:
src = butler.get('src', dataId={'filter': 'HSC-R', 'visit': 903334, 'ccd': 23})
This src dataset is a SourceTable, which is a table object from the lsst.afw.table module.
You’ll explore SourceTables more in a later tutorial, but you can check its length with Python’s len function:
print(len(src))
The columns of a table are defined in its schema. You can print out the schema to see each column’s name, data type, and description:
print(src.getSchema())
To get just the names of columns, run:
print(src.getSchema().getNames())
To get metadata about a specific column, like calib_pdfUsed:
print(src.schema.find("calib_psfUsed"))
Given a name, you can get a column’s values as a familiar Numpy array like this:
print(src['base_PsfFlux_flux'])
Tip
If you are working in a Jupyter notebook you can see an HTML table rendering of any lsst.afw.table table object by getting an astropy.table.Table version of it:
src.asAstropy()
The returned Astropy Table is a view, not a copy, so it doesn’t consume much additional memory.
Plotting sources on the display¶
Now you’ll overplot sources from the src table onto the image display using the Display’s dot method for plotting markers.
Display.dot plots markers individually, so you’ll need to iterate over rows in the SourceTable.
It’s more efficient to send a batch of updates to the display, though, so enclose the loop in a display.Buffering context, like this:
with display.Buffering():
for s in src:
display.dot("o", s.getX(), s.getY(), size=10, ctype='orange')
Now orange circles should appear in the DS9 window over every detected source.
Note
Notice the getX and getY methods for getting the (x,y) centroid of each source.
These methods are shortcuts, using the table’s slot system.
Because the the src catalog contains measurements from several measurement plugins, slots are a way of easily using the pre-configured best measurements of a source.
Clearing markers¶
Display.dot always adds new markers to the display.
To clear the display of all markers, use the erase method:
display.erase()
Selecting PSF-fitting sources to plot on the display¶
Next, use the display to understand what sources were used for PSF measurement.
The src table’s calib_psfUsed column describes whether the source was used for PSF measurement.
Since columns are Numpy arrays we can iterate over rows where src['calib_psfUsed'] is True with Numpy’s boolean array indexing:
with display.Buffering():
for s in src[src['calib_psfUsed']]:
display.dot("x", s.getX(), s.getY(), size=10, ctype='red')
Red x symbols on the display mark all stars used by PSF measurement.
Some sources might be considered as PSF candidates, but later rejected.
In this statement, you can use a logical & (and) operator to combine boolean index arrays where both src['calib_psfCandidate'] is True and src['calib_psfUsed'] == False as well:
rejectedPsfSources = src[src['calib_psfCandidate'] &
(src['calib_psfUsed'] == False)]
with display.Buffering():
for s in rejectedPsfSources:
display.dot("+", s.getX(), s.getY(), size=10, ctype='green')
Now all green plus (+) symbols on the display mark rejected PSF measurement sources.
The display framework, as you’ve seen, is a useful facility for inspecting images and tables. This tutorial only covered the framework’s basic functionality. Explore the display framework documentation to learn how to display multiple images at once, and to work with different display backends.
Wrap up¶
In this tutorial you’ve worked with the LSST Science Pipelines Python API to display images and tables. Here are some key takeaways:
- Use the
lsst.daf.persistence.Butlerclass to read and write data from repositories. - The
lsst.afw.displaymodule provides a flexible framework for sending data from LSST Science Pipelines code to image displays. You used the DS9 backend in this tutorial, but other backends are available. - Exposure objects have image data, mask data, and metadata. When you display an exposure, the display framework automatically overlays mask planes.
- Tables have well-defined schemas. Use methods like
getSchemato understand the contents of a table. You can also use theasAstropytable method to view the table as anastropy.table.Table.
Continue this tutorial series in part 4, where you’ll coadd these processed images into deeper mosaics.